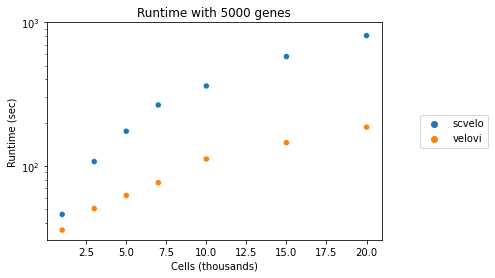
VeloVI Runtime Analysis#
Download Data#
Using data from pachterlab/MBLGLMBHGP_2021.
!wget -O retina.h5ad.gz https://caltech.box.com/shared/static/lo6satyyrjhvhgkmj3gv39360eshxa3n.gz
!gunzip retina.h5ad.gz
Imports#
import time
import random
import itertools
import os
import anndata
import numpy as np
import pandas as pd
from sklearn.preprocessing import MinMaxScaler
import torch
import scanpy as sc
import scvelo as scv
from velovi import VELOVI
Global seed set to 0
Helper Definitions#
def fit_scvelo(adata):
start = time.time()
scv.tl.recover_dynamics(
adata, fit_scaling=True, var_names=adata.var_names, n_jobs=8
)
scv.tl.velocity(adata, mode="dynamical")
return time.time() - start
def fit_velovi(bdata):
start = time.time()
VELOVI.setup_anndata(bdata, spliced_layer="Ms", unspliced_layer="Mu")
vae = VELOVI(bdata)
vae.train()
velocities = vae.get_velocity(n_samples=25, velo_statistic="mean")
return time.time() - start
def min_max_scaled(bdata):
mu_scaler = MinMaxScaler()
bdata.layers['Mu'] = mu_scaler.fit_transform(bdata.layers['Mu'])
ms_scaler = MinMaxScaler()
bdata.layers['Ms'] = ms_scaler.fit_transform(bdata.layers['Ms'])
return bdata
Run tests#
adata = anndata.read_h5ad("retina.h5ad")
adata
AnnData object with n_obs × n_vars = 113909 × 5000
obs: 'sample', 'time', 'barcode', 'umap_cluster', 'umap_coord1', 'umap_coord2', 'umap_coord3', 'used_for_pseudotime', 'umap2_CellType', 'n_genes', 'initial_size_spliced', 'initial_size_unspliced', 'initial_size', 'n_counts', 'velocity_self_transition'
var: 'n_cells', 'means', 'dispersions', 'dispersions_norm', 'highly_variable', 'velocity_gamma', 'velocity_r2', 'velocity_genes'
uns: 'neighbors', 'pca', 'umap2_CellType_colors', 'velocity_graph', 'velocity_graph_neg', 'velocity_params'
obsm: 'X_pca', 'X_umap', 'velocity_pca', 'velocity_umap'
varm: 'PCs'
layers: 'Ms', 'Mu', 'spliced', 'unspliced', 'variance_velocity', 'velocity'
obsp: 'connectivities', 'distances'
def run_runtime_tests(adata, subsample_ns):
methods = ["scvelo", "velovi"]
configs = list(itertools.product(methods, subsample_ns))
random.shuffle(configs)
for method_name, subsample_n in configs:
save_name = f"results/{method_name}_{subsample_n}"
if os.path.exists(f"{save_name}.npy"):
print(f"{save_name} already exists, skipping")
continue
print(f"Running {method_name} with subsample_n={subsample_n}.")
bdata = sc.pp.subsample(adata, n_obs=subsample_n, copy=True)
if method_name == "scvelo":
runtime_s = fit_scvelo(bdata)
elif method_name == "velovi":
bdata = min_max_scaled(bdata)
runtime_s = fit_velovi(bdata)
else:
raise ValueError
res_row = [subsample_n, runtime_s, method_name]
np.save(save_name, np.array(res_row))
run_runtime_tests(adata, subsample_ns=[1000, 3000, 5000, 7500, 10000, 15000, 20000])
results/scvelo_3000 already exists, skipping
results/velovi_7500 already exists, skipping
results/velovi_5000 already exists, skipping
Running scvelo with subsample_n=15000.
recovering dynamics (using 8/20 cores)
WARNING: ENSMUSG00000047562 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000025328 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000071561 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000016982 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000006179 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000031932 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000061100 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000028946 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000059762 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000054986 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000100426 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000085035 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000040610 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000043230 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000049685 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000052974 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000046480 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000026308 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000059412 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000105105 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000105352 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000041559 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000106136 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000031506 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000097386 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000094891 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000041287
WARNING: ENSMUSG00000032346 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000036594 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000042485 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000054406 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000109485 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000025105 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000035187 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000028071 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000029307 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000029304 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000026581 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000040629
WARNING: ENSMUSG00000022220 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000001020 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000053318 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000101746 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000009350 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000001508 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000056648 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000097850 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000038216 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000051457 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000042985 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000031138 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000056380 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000079845 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000038745 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000001494
WARNING: ENSMUSG00000071229 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000018486 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000053129 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000026805 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000108961 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000031449 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000006570 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000042386 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000059201 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000087613 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000024619 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000000394 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000050473 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000033006 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000074240 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000085404 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000074217 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000009654
WARNING: ENSMUSG00000086905 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000059654 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000034783 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000022991 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000075023 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000025418 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000034785 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000028610 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000023039 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000064232 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000061397 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000024678 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000035963 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000020125 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000039209 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000031722 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000021095 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000097558 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000074896 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000062488 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000041347 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000074805 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000002803 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000024979 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000090965 not recoverable due to insufficient samples.
finished (0:09:10) --> added
'fit_pars', fitted parameters for splicing dynamics (adata.var)
computing velocities
finished (0:00:26) --> added
'velocity', velocity vectors for each individual cell (adata.layers)
results/velovi_10000 already exists, skipping
results/scvelo_5000 already exists, skipping
Running scvelo with subsample_n=7500.
recovering dynamics (using 8/20 cores)
WARNING: skipping bimodality check for ENSMUSG00000032769
WARNING: ENSMUSG00000047562 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000025328 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000071561 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000016982 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000006179 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000074695 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000031932 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000030156 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000053063 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000058550 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000061100 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000028946 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000059762 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000054986 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000078949
WARNING: ENSMUSG00000100426 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000078810
WARNING: ENSMUSG00000012428 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000085035 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000021469 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000006542 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000047109 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000043230 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000040610 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000011349 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000046480 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000049685 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000052974 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000059412 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000104000 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000026308 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000055546 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000032315 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000001865 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000059956 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000064147
WARNING: ENSMUSG00000105105 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000105352 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000041559 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000031506 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000030470 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000106136 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000094891 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000032346 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000049511 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000027833 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000037548 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000036594 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000097386 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000034923 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000041287 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000042485 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000006522 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000028071 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000109485 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000025105 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000038540 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000054406 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000035187 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000029307 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000029306 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000029304 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000023968 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000026581 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000040629 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000023993 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000001020 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000044037
WARNING: ENSMUSG00000053318 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000024164
WARNING: ENSMUSG00000022220 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000015443 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000073907 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000101746 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000027871 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000089842
WARNING: ENSMUSG00000009350 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000072553 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000097850 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000046213 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000022097 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000030762 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000001508 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000056648 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000051457 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000030862
WARNING: ENSMUSG00000038216 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000042985 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000038745 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000075570 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000031138 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000056380 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000079845 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000001494 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000028270 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000098630 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000106052 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000026805 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000071229 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000108888 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000018486 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000108961 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000053129 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000031449 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000006570 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000044743 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000073008
WARNING: ENSMUSG00000042386 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000029755 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000086644
WARNING: ENSMUSG00000024619 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000059201 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000000394 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000087613 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000050473 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000037440 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000085404 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000043505 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000033006 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000086905 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000020591 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000074240 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000074217 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000009654 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000075023 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000025418 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000034785 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000028610 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000024677 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000024678 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000098854 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000055895 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000059654 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000022991 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000034783 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000034226 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000023039 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000064232 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000061397 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000035963 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000097558 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000074896 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000062488 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000020125 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000033498 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000031722 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000050234 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000074805 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000039209 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000050108
WARNING: ENSMUSG00000028778 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000021095 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000041347 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000019936 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000024979 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000035818 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000090965 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000002803 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000004366 not recoverable due to insufficient samples.
finished (0:04:12) --> added
'fit_pars', fitted parameters for splicing dynamics (adata.var)
computing velocities
finished (0:00:13) --> added
'velocity', velocity vectors for each individual cell (adata.layers)
Running velovi with subsample_n=1000.
GPU available: True, used: True
TPU available: False, using: 0 TPU cores
IPU available: False, using: 0 IPUs
HPU available: False, using: 0 HPUs
LOCAL_RANK: 0 - CUDA_VISIBLE_DEVICES: [0]
Epoch 361/500: 72%|███████▏ | 361/500 [00:33<00:12, 10.76it/s, loss=-1.92e+04, v_num=1]
Monitored metric elbo_validation did not improve in the last 45 records. Best score: -15787.076. Signaling Trainer to stop.
Running velovi with subsample_n=3000.
GPU available: True, used: True
TPU available: False, using: 0 TPU cores
IPU available: False, using: 0 IPUs
HPU available: False, using: 0 HPUs
LOCAL_RANK: 0 - CUDA_VISIBLE_DEVICES: [0]
Epoch 192/500: 38%|███▊ | 192/500 [00:47<01:15, 4.07it/s, loss=-2.11e+04, v_num=1]
Monitored metric elbo_validation did not improve in the last 45 records. Best score: -19836.871. Signaling Trainer to stop.
Running scvelo with subsample_n=10000.
recovering dynamics (using 8/20 cores)
WARNING: ENSMUSG00000047562 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000025328 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000031932 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000016982 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000006179 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000071561 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000074695 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000030156 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000053063 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000058550 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000061100 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000028946 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000059762 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000054986 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000100426 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000085035 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000047109 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000040610 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000011349 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000049685 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000052974 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000046480 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000043230 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000059412 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000001865 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000026308 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000105105 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000105352 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000059956 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000106136 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000094891 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000030470 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000027833 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000037548 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000036594 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000034923 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000097386 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000032346 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000041559 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000031506 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000041287 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000035187 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000042485 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000028071 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000029307 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000029306 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000029304 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000109485 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000025105 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000038540
WARNING: ENSMUSG00000023968 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000054406 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000023993 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000001020 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000026581 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000040629 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000073907 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000022220 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000053318 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000027871
WARNING: ENSMUSG00000101746 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000009350 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000030762
WARNING: ENSMUSG00000072553 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000042985 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000051457 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000022097 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000097850 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000001508 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000056648 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000031138 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000038216 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000038745 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000056380 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000079845 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000028270 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000075570 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000053129 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000001494
WARNING: ENSMUSG00000108888 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000108961 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000098630
WARNING: ENSMUSG00000029755 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000031449 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000071229 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000006570 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000018486 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000026805 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000042386 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000059201 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000087613 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000024619 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000043505 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000000394 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000050473 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000085404 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000074240 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000074217 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000059654 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000009654
WARNING: ENSMUSG00000034783 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000098854 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000025418 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000034785 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000028610 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000033006 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000020591 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000024677 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000024678 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000055895
WARNING: ENSMUSG00000086905 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000075023 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000022991 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000031722 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000035963 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000020125 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000097558 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000074896 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000062488 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000050234
WARNING: ENSMUSG00000023039 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000064232 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000061397 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000028778 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000034226 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000039209 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000024979 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000035818
WARNING: ENSMUSG00000090965 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000021095 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000074805 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000041347 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000002803 not recoverable due to insufficient samples.
finished (0:05:42) --> added
'fit_pars', fitted parameters for splicing dynamics (adata.var)
computing velocities
finished (0:00:18) --> added
'velocity', velocity vectors for each individual cell (adata.layers)
Running scvelo with subsample_n=1000.
recovering dynamics (using 8/20 cores)
WARNING: skipping bimodality check for ENSMUSG00000025902
WARNING: ENSMUSG00000041886 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000086146 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000074735 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000079008 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000068129 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000097312 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000101476 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000032769 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000047976 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000025888 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000033538 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000030345
WARNING: ENSMUSG00000047562 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000069516 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000100053 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000097525
WARNING: ENSMUSG00000074695 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000025328 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000087045 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000034362 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000071561 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000027470 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000053101 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000016982 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000006179 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000047228 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000031932 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000030156 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000053063 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000030159 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000033082 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000108763 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000021336 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000033770 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000025432 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000038402 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000042842 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000027611 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000022667 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000042638 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000030218 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000039109
WARNING: ENSMUSG00000022659 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000040345 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000006235 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000058550 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000061100 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000104072 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000087094 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000078949 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000028946 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000085649 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000059762 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000087277 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000048776 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000054986 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000098590 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000058755 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000042116 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000034201 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000029070 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000078810 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000032125 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000001943 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000087684 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000051095 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000100426 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000073234 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000012428 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000101895 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000070870 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000044429 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000021469 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000021478 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000002033 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000046480 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000059412 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000047109 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000085035 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000039155 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000006542 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000033227 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000026205 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000049625 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000095550 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000035493 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000002633 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000025746 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000026202 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000067642 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000026208 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000040899 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000029163 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000021456 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000023828 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000043230 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000060969 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000023885 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000097566 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000045587 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000011349 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000003379 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000026222 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000040610 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000032911 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000043747 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000026241 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000026247
WARNING: ENSMUSG00000049685 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000052974 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000104000 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000086719 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000048992 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000032315 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000104015 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000044716 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000034486 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000026303 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000026304 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000046491 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000060913 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000026308 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000001865 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000097809
WARNING: ENSMUSG00000074768 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000034107 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000062753 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000055546 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000062132 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000100182 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000074199 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000009687
WARNING: ENSMUSG00000021678 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000020377 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000050087 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000059956 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000029092 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000001053 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000064147 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000052485 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000032204
WARNING: ENSMUSG00000105105 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000105352 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000071203 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000024029 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000020264 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000074604 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000004609 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000064023 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000027744 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000105168 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000106270 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000052142 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000027750 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000068082 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000094891 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000002289 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000085551 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000041559 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000106136 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000037548 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000036594 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000032346 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000030792 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000031506 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000027833 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000049511 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000034923 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000041801 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000032265 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000060180 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000026413
WARNING: ENSMUSG00000009487 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000072845 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000054537 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000029268 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000106677 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000029272 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000048304
WARNING: ENSMUSG00000030470 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000099974 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000089929
WARNING: ENSMUSG00000039518 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000032356 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000105640 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000097386 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000032807 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000029370 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000029373 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000026357 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000029378 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000041287 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000090897 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000108313 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000089887 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000106357 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000100123 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000028071 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000105218 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000004892 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000042641 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000100627 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000035187 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000023968 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000067144 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000042485 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000006522 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000085230 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000029307 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000029306 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000029304 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000106483 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000047150 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000039236 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000030607 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000029275 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000023992 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000023993 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000005950 not recoverable due to insufficient samples.
WARNING: WARNING:ENSMUSG00000041707 not recoverable due to insufficient samples.ENSMUSG00000010592
not recoverable due to insufficient samples.
WARNING: ENSMUSG00000026581 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000054406 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000066975 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000021799 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000040629 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000109485 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000025105 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000001020 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000038540 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000027913 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000054675 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000032491 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000044037 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000024164 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000013707 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000097045 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000015355
WARNING: ENSMUSG00000029564 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000015316 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000017390 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000060523 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000028108 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000035932 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000007097 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000032530 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000044534 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000041548
WARNING: ENSMUSG00000053318 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000030732 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000025804 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000030725 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000086557 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000074388 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000031147
WARNING: ENSMUSG00000035373 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000106708 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000066671 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000082101 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000027871 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000086564 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000031165 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000031170 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000022212 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000019122 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000032725 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000101746 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000022220 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000015443 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000026497 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000018698 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000002228 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000040505 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000070421 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000073907 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000037060 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000090086 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000074342 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000091649 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000039377 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000040724 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000046213 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000089842 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000036557 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000039349
WARNING: skipping bimodality check for ENSMUSG00000030787
WARNING: ENSMUSG00000051827 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000079638 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000095741 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000009350 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000031101 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000073862 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000072553 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000057072
WARNING: ENSMUSG00000021966 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000030666 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000061808 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000108856 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000060967 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000031125 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000087344 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000031138 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000001506 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000001508 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000096946 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000097850 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000056380 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000016494 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000026645 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000079845 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000046719 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000004814 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000031375
WARNING: ENSMUSG00000002007 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000056648 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000042985 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000030762 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000085257 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000022097 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000033589 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000019232 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000034997 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000036949 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000038216 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000051457 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000025058 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000030789 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000035831 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000075570 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000073599 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000099422 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000050505 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000030862 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000044628 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000100717 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000026956 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000028270 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000024440 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000031297 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000036586
WARNING: ENSMUSG00000106052 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000004044 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000093910 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000038745 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000025491 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000025499
WARNING: skipping bimodality check for ENSMUSG00000052821
WARNING: ENSMUSG00000060240 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000073010 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000024503 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000054293
WARNING: ENSMUSG00000098630 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000073008 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000001494 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000028037 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000028036 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000000244 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000071229 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000029641 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000053129 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000026805 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000109009 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000028174
WARNING: ENSMUSG00000108888 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000033436
WARNING: ENSMUSG00000018486 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000108961 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000109508 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000096938 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000024610 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000042386 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000003882 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000024619 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000022243 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000010435 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000031449 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000002588 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000067276 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000020712 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000001029
WARNING: ENSMUSG00000052951 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000019577 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000029755 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000061762 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000006570 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000044743 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000025333 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000026882 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000020614 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000028298
WARNING: ENSMUSG00000031549 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000037406 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000048230 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000087137 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000086644 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000031489 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000059201 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000086523 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000036078 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000051517 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000084861 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000026832 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000097117 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000087613 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000026971 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000025044 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000051225 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000000394 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000053469 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000019851 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000029915 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000050473 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000037440 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000045903 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000045281 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000039691 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000031610 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000024910 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000054435 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000043505 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000085404 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000029831 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000097742 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000053886 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000005716 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000071715 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000071711
WARNING: ENSMUSG00000000938 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000051675 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000063428 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000033220 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000018008 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000004655 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000037984 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000033006 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000019909 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000097580
WARNING: ENSMUSG00000020591 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000000489 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000063796 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000027077 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000046718 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000009654 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000086905 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000020641 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000053977 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000074240 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000036136 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000108371 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000020651 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000020096 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000024672 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000024675 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000079419
WARNING: ENSMUSG00000024677 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000024678 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000024679 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000055895 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000027221 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000020545
WARNING: ENSMUSG00000031710 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000075023 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000025418 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000074217 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000034785 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000028610 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000059654 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000058624 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000016458 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000045608 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000034783 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000028716 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000086896
WARNING: ENSMUSG00000028713 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000020178 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000098854 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000063415 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000048572 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000000903 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000042372 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000022991 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000001155 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000000290 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000068614 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000023484 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000015053 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000035963 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000020310 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000089671 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000056973 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000024778 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000097558 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000074896 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000062488 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000047203 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000020125 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000031760 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000034226 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000027313 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000023039 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000064232 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000061397 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000074128 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000033498 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000033249 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000021032 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000031891
WARNING: ENSMUSG00000044287 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000021886 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000022516
WARNING: ENSMUSG00000074852 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000039457 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000051669 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000027360
WARNING: ENSMUSG00000031722 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000050234 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000039209 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000022501 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000050108 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000028786 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000000440 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000020051 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000064215 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000079017 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000021091 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000021095 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000020062 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000047638 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000028778 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000028581 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000041347 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000025701 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000027398 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000074805 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000019936 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000074776 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000022738 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000024979 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000056912
WARNING: ENSMUSG00000035818 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000019890
WARNING: ENSMUSG00000027345 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000036602 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000030109 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000030116 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000027261 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000084762 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000090965 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000030148 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000095486 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000008845 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000002803 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000004366 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000056423 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000036905 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000036896 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000036887 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000047953 not recoverable due to insufficient samples.
finished (0:00:44) --> added
'fit_pars', fitted parameters for splicing dynamics (adata.var)
computing velocities
finished (0:00:01) --> added
'velocity', velocity vectors for each individual cell (adata.layers)
Running velovi with subsample_n=15000.
GPU available: True, used: True
TPU available: False, using: 0 TPU cores
IPU available: False, using: 0 IPUs
HPU available: False, using: 0 HPUs
LOCAL_RANK: 0 - CUDA_VISIBLE_DEVICES: [0]
Epoch 112/500: 22%|██▏ | 112/500 [02:09<07:28, 1.16s/it, loss=-2.34e+04, v_num=1]
Monitored metric elbo_validation did not improve in the last 45 records. Best score: -22225.021. Signaling Trainer to stop.
Running scvelo with subsample_n=20000.
recovering dynamics (using 8/20 cores)
WARNING: ENSMUSG00000047562 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000071561 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000016982 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000006179 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000028946 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000061100 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000059762 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000054986 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000100426 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000085035 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000040610 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000046480 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000049685 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000052974 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000059412 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000026308 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000105105 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000105352 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000106136 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000041559 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000094891
WARNING: ENSMUSG00000031506 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000097386 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000036594 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000028071 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000042485 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000035187 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000025105 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000054406 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000029307 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000029304 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000001020 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000026581 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000022220 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000053318 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000101746 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000009350 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000051457 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000042985 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000001508 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000097850 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000056648
WARNING: ENSMUSG00000031138 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000056380 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000079845 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000038745 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000038216 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000053129 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000071229 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000001494
WARNING: ENSMUSG00000108961 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000031449 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000006570 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000018486 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000042386 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000059201 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000024619 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000050473 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000000394 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000074240 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000085404 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000074217 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000059654 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000033006 not recoverable due to insufficient samples.
WARNING: skipping bimodality check for ENSMUSG00000009654
WARNING: ENSMUSG00000034783 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000025418 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000034785 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000086905 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000022991 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000075023 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000035963 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000020125 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000023039 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000064232 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000061397 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000024678 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000031722 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000039209 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000074896 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000062488 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000021095 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000074805 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000041347 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000002803 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000024979 not recoverable due to insufficient samples.
WARNING: ENSMUSG00000090965 not recoverable due to insufficient samples.
finished (0:12:41) --> added
'fit_pars', fitted parameters for splicing dynamics (adata.var)
computing velocities
finished (0:00:46) --> added
'velocity', velocity vectors for each individual cell (adata.layers)
results/velovi_20000 already exists, skipping
Plotting#
import glob
runtime_result_files = glob.glob("results/**.npy")
res_df_rows = []
for res_file in runtime_result_files:
res_df_row = np.load(res_file)
res_df_rows.append(res_df_row)
res_df = pd.DataFrame(res_df_rows, columns=['n_obs', 'runtime', 'method'])
import matplotlib.pyplot as plt
import seaborn as sns
import mplscience
res_df['n_obs_thousand'] = res_df['n_obs'].astype(int) // 1e3
res_df['runtime'] = res_df['runtime'].astype(float)
fig, ax = plt.subplots(figsize=(6, 4))
with mplscience.style_context():
sns.scatterplot(x='n_obs_thousand', y='runtime', hue='method', data=res_df, ax=ax)
ax.set_xlabel('Cells (thousands)')
ax.set_ylabel('Runtime (sec)')
ax.set_yscale('log')
ax.set_ylim((0, 1e3))
ax.set_title('Runtime with 5000 genes')
ax.legend(loc='center left', bbox_to_anchor=(1.1, 0.5))
# plt.show()
plt.savefig('figures/retina_runtime.svg', bbox_inches='tight', format="svg")