RNA velocity in adult mouse brain#
RNA velocity analysis with the VI model using data preprocessed with alevin_spliced_unspliced_gentrome.
Requires
adata_generation.ipynbvelocyto_var_names.csvfromvelocyto_vi.ipynb
Output
DATA_DIR/old_brain/velocities/alevin_spliced_unspliced_velovi.npy
Library imports#
import os
from pathlib import Path
import sys
import numpy as np
import pandas as pd
import torch
from sklearn.preprocessing import MinMaxScaler
import matplotlib.pyplot as plt
import mplscience
import seaborn as sns
import scanpy as sc
import scvelo as scv
import scvi
from velovi import VELOVI
sys.path.insert(0, "../../../")
from paths import DATA_DIR
Global seed set to 0
sc.logging.print_version_and_date()
Running Scanpy 1.9.1, on 2022-07-19 21:25.
General settings#
scvi.settings.dl_pin_memory_gpu_training = False
# set verbosity levels
sc.settings.verbosity = 2
scv.settings.verbosity = 3
scv.settings.set_figure_params('scvelo', dpi_save=400, dpi=80, transparent=True, fontsize=20, color_map='viridis')
scv.settings.plot_prefix = ""
Constants#
VELOCYTO_VAR_NAMES = pd.read_csv('velocyto_var_names.csv', index_col=0, header=None).index.tolist()
Function definitions#
def fit_velovi(bdata):
VELOVI.setup_anndata(bdata, spliced_layer="Ms", unspliced_layer="Mu")
vae = VELOVI(bdata)
vae.train()
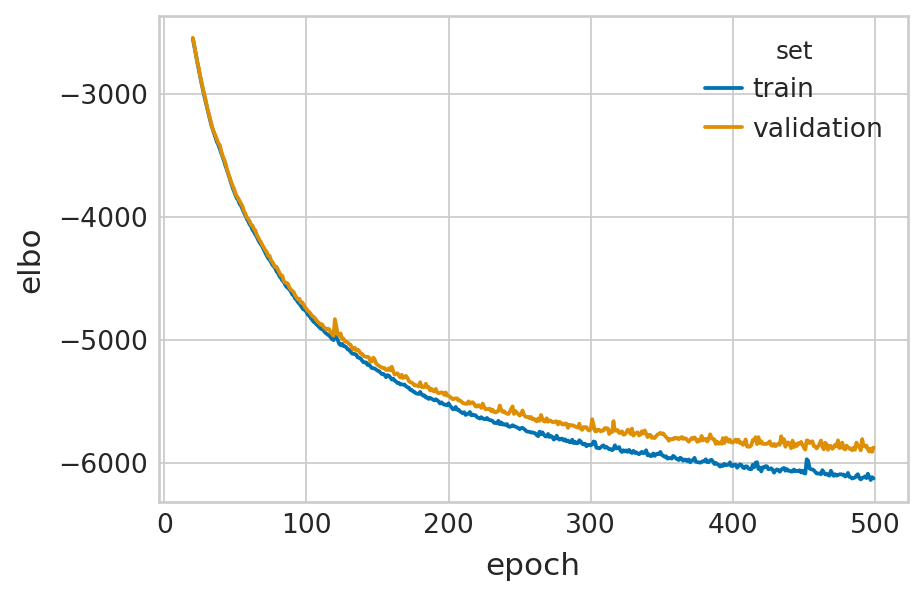
df = vae.history["elbo_train"].iloc[20:].reset_index().rename(columns={'elbo_train': 'elbo'})
df['set'] = 'train'
_df = vae.history["elbo_validation"].iloc[20:].reset_index().rename(columns={'elbo_validation': 'elbo'})
_df['set'] = 'validation'
df = pd.concat([df, _df], axis=0).reset_index(drop=True)
with mplscience.style_context():
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(figsize=(6, 4))
sns.lineplot(data=df, x='epoch', y='elbo', hue='set', palette=['#0173B2', '#DE8F05'], ax=ax)
latent_time = vae.get_latent_time(n_samples=25)
velocities = vae.get_velocity(n_samples=25, velo_statistic="mean")
t = latent_time
scaling = 20 / t.max(0)
bdata.layers["velocities_velovi"] = velocities / scaling
bdata.layers["latent_time_velovi"] = latent_time
bdata.var["fit_alpha"] = vae.get_rates()["alpha"] / scaling
bdata.var["fit_beta"] = vae.get_rates()["beta"] / scaling
bdata.var["fit_gamma"] = vae.get_rates()["gamma"] / scaling
bdata.var["fit_t_"] = (
torch.nn.functional.softplus(vae.module.switch_time_unconstr)
.detach()
.cpu()
.numpy()
) * scaling
bdata.layers["fit_t"] = latent_time.values * scaling[np.newaxis, :]
bdata.var['fit_scaling'] = 1.0
return vae
Data loading#
adata = sc.read(DATA_DIR / 'old_brain' / "alevin_spliced_unspliced_gentrome.h5ad")
adata
AnnData object with n_obs × n_vars = 1823 × 54143
obs: 'cell_index', 'clusters_coarse', 'clusters', 'NAME', 'nGene', 'nUMI', 'animal_type', 'cell_type_age'
layers: 'spliced', 'unspliced'
scv.pl.proportions(adata)
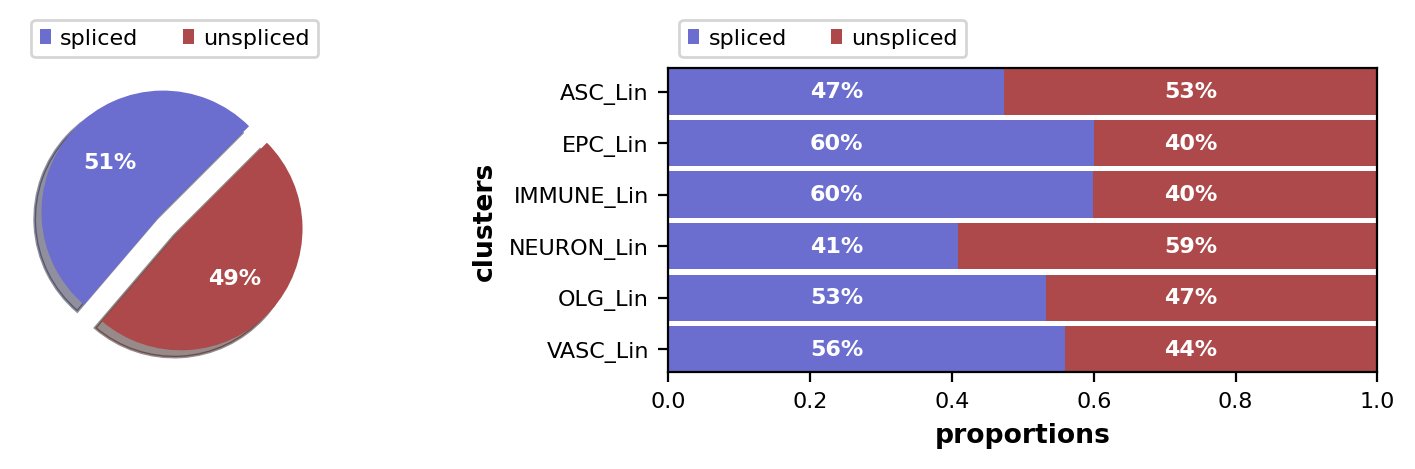
Data pre-processing#
scv.pp.filter_and_normalize(adata, min_shared_counts=20, n_top_genes=2000, retain_genes=VELOCYTO_VAR_NAMES)
scv.pp.moments(adata, n_pcs=30, n_neighbors=30)
Filtered out 41946 genes that are detected 20 counts (shared).
WARNING: Did not normalize X as it looks processed already. To enforce normalization, set `enforce=True`.
WARNING: Did not normalize spliced as it looks processed already. To enforce normalization, set `enforce=True`.
WARNING: Did not normalize unspliced as it looks processed already. To enforce normalization, set `enforce=True`.
Extracted 2816 highly variable genes.
Logarithmized X.
computing PCA
on highly variable genes
with n_comps=30
finished (0:00:02)
computing neighbors
finished (0:00:36) --> added
'distances' and 'connectivities', weighted adjacency matrices (adata.obsp)
computing moments based on connectivities
finished (0:00:04) --> added
'Ms' and 'Mu', moments of un/spliced abundances (adata.layers)
mu_scaler = MinMaxScaler()
adata.layers["Mu"] = mu_scaler.fit_transform(adata.layers["Mu"])
ms_scaler = MinMaxScaler()
adata.layers["Ms"] = ms_scaler.fit_transform(adata.layers["Ms"])
scv.tl.velocity(adata, mode="steady_state")
adata = adata[:, VELOCYTO_VAR_NAMES].copy()
computing velocities
finished (0:00:01) --> added
'velocity', velocity vectors for each individual cell (adata.layers)
Model fitting#
fit_velovi(adata)
/home/icb/philipp.weiler/miniconda3/envs/velovi-py39/lib/python3.9/site-packages/torch/distributed/_sharded_tensor/__init__.py:8: DeprecationWarning: torch.distributed._sharded_tensor will be deprecated, use torch.distributed._shard.sharded_tensor instead
warnings.warn(
GPU available: True, used: True
TPU available: False, using: 0 TPU cores
IPU available: False, using: 0 IPUs
LOCAL_RANK: 0 - CUDA_VISIBLE_DEVICES: [0]
Set SLURM handle signals.
Epoch 500/500: 100%|██████████| 500/500 [04:20<00:00, 1.92it/s, loss=-6.12e+03, v_num=1]
VELOVI Model with the following params: n_hidden: 256, n_latent: 10, n_layers: 1, dropout_rate: 0.1 Training status: Trained
Save results#
np.save(
file=DATA_DIR / 'old_brain' / 'velocities' / 'alevin_spliced_unspliced_velovi.npy', arr=adata.layers['velocities_velovi']
)