Comparison of inferred velocity and latent times#
Requires
fitted_values.csvgenerated bypancreas.ipynb
Library imports#
import os
import sys
import numpy as np
import pandas as pd
from scipy.stats import pearsonr, spearmanr, ttest_ind
from sklearn.preprocessing import MinMaxScaler
import matplotlib.pyplot as plt
import mplscience
import seaborn as sns
import scvi
import scvelo as scv
import torch
from velovi import VELOVI
sys.path.append("../..")
from utils._simulation import simulate_data
from paths import DATA_DIR, FIG_DIR
Global seed set to 0
General settings#
scvi.settings.dl_pin_memory_gpu_training = False
sns.reset_defaults()
sns.reset_orig()
scv.settings.set_figure_params('scvelo', dpi_save=400, dpi=80, transparent=True, fontsize=20, color_map='viridis')
SAVE_FIGURES = True
if SAVE_FIGURES:
os.makedirs(FIG_DIR / 'comparison', exist_ok=True)
Function definition#
def fit_scvelo(adata):
scv.tl.recover_dynamics(
adata, var_names=adata.var_names, use_raw=True, n_jobs=8
)
scv.tl.velocity(adata, mode="dynamical")
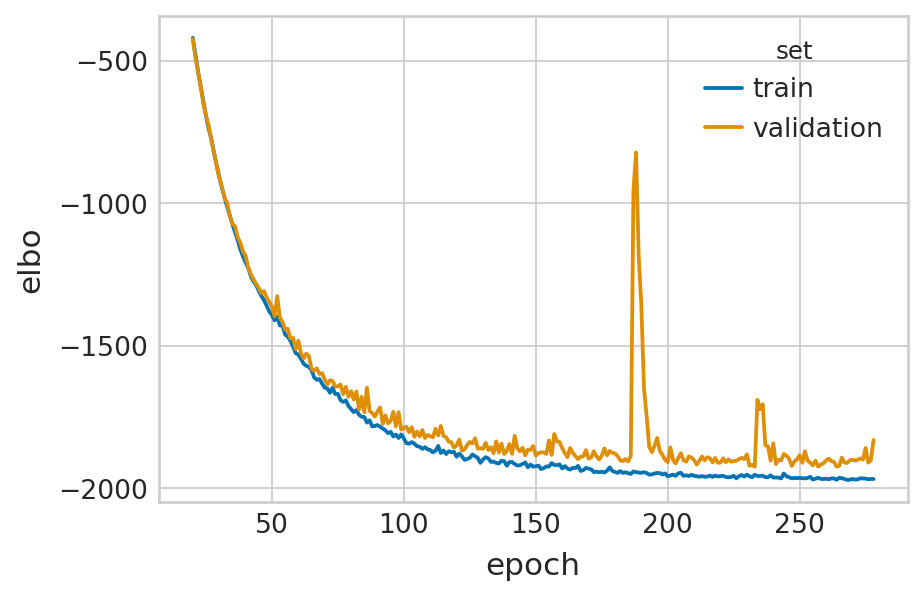
def fit_velovi(bdata):
VELOVI.setup_anndata(bdata, spliced_layer="Ms", unspliced_layer="Mu")
vae = VELOVI(bdata)
vae.train()
df = vae.history["elbo_train"].iloc[20:].reset_index().rename(columns={'elbo_train': 'elbo'})
df['set'] = 'train'
_df = vae.history["elbo_validation"].iloc[20:].reset_index().rename(columns={'elbo_validation': 'elbo'})
_df['set'] = 'validation'
df = pd.concat([df, _df], axis=0).reset_index(drop=True)
with mplscience.style_context():
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(figsize=(6, 4))
sns.lineplot(data=df, x='epoch', y='elbo', hue='set', palette=['#0173B2', '#DE8F05'], ax=ax)
latent_time = vae.get_latent_time(n_samples=25)
velocities = vae.get_velocity(n_samples=25, velo_statistic="mean")
t = latent_time
scaling = 20 / t.max(0)
bdata.layers["velocity"] = velocities / scaling
bdata.layers["latent_time"] = latent_time
bdata.var["fit_alpha"] = vae.get_rates()["alpha"] / scaling
bdata.var["fit_beta"] = vae.get_rates()["beta"] / scaling
bdata.var["fit_gamma"] = vae.get_rates()["gamma"] / scaling
bdata.var["fit_t_"] = (
torch.nn.functional.softplus(vae.module.switch_time_unconstr)
.detach()
.cpu()
.numpy()
) * scaling
bdata.layers["fit_t"] = latent_time.values * scaling[np.newaxis, :]
bdata.var['fit_scaling'] = 1.0
return vae
Data loading#
df = pd.read_csv(DATA_DIR / "pancreas" / "fitted_values.csv", index_col=0).sample(frac=0.5)
df.head()
| fit_alpha | fit_beta | fit_gamma | fit_t_ | model | |
|---|---|---|---|---|---|
| index | |||||
| Nptx2 | 0.083103 | 0.108307 | 0.188514 | 14.631201 | VI |
| Rbpjl | 0.026645 | 0.337411 | 0.206346 | 21.273163 | VI |
| Syt7 | 0.148926 | 0.282632 | 0.205117 | 17.131525 | EM |
| Cdc14b | 0.158472 | 0.277238 | 0.203630 | 19.300320 | EM |
| Cxxc4 | 0.127181 | 0.183402 | 0.267991 | 12.156104 | VI |
adata = simulate_data(
alpha=df['fit_alpha'].values,
beta=df['fit_beta'].values,
gamma=df['fit_gamma'].values,
t_switch=df['fit_t_'].values,
)
adata.var['model'] = df['model'].values
adata
AnnData object with n_obs × n_vars = 3000 × 1074
obs: 'true_t'
var: 'true_t_', 'true_alpha', 'true_beta', 'true_gamma', 'true_scaling', 'model'
layers: 'unspliced', 'spliced', 'true_velocity'
adata.layers['Mu'] = adata.layers['unspliced']
adata.layers['Ms'] = adata.layers['spliced']
bdata = adata.copy()
Parameter inference#
EM model#
fit_scvelo(adata)
recovering dynamics (using 8/14 cores)
Global seed set to 0
Global seed set to 0
Global seed set to 0
Global seed set to 0
Global seed set to 0
Global seed set to 0
Global seed set to 0
Global seed set to 0
finished (0:00:26) --> added
'fit_pars', fitted parameters for splicing dynamics (adata.var)
computing velocities
finished (0:00:01) --> added
'velocity', velocity vectors for each individual cell (adata.layers)
scv.pl.scatter(adata, basis=adata.var_names[:20], ncols=5, frameon=False)

VI model#
velovi_vae = fit_velovi(bdata)
GPU available: True (cuda), used: True
TPU available: False, using: 0 TPU cores
IPU available: False, using: 0 IPUs
HPU available: False, using: 0 HPUs
LOCAL_RANK: 0 - CUDA_VISIBLE_DEVICES: [0]
Epoch 279/500: 56%|█████▌ | 279/500 [02:00<01:35, 2.31it/s, loss=-1.97e+03, v_num=1]
Monitored metric elbo_validation did not improve in the last 45 records. Best score: -1923.831. Signaling Trainer to stop.
scv.pl.scatter(bdata, basis=bdata.var_names[:20], ncols=5, frameon=False)
Correlation analysis#
Velocity#
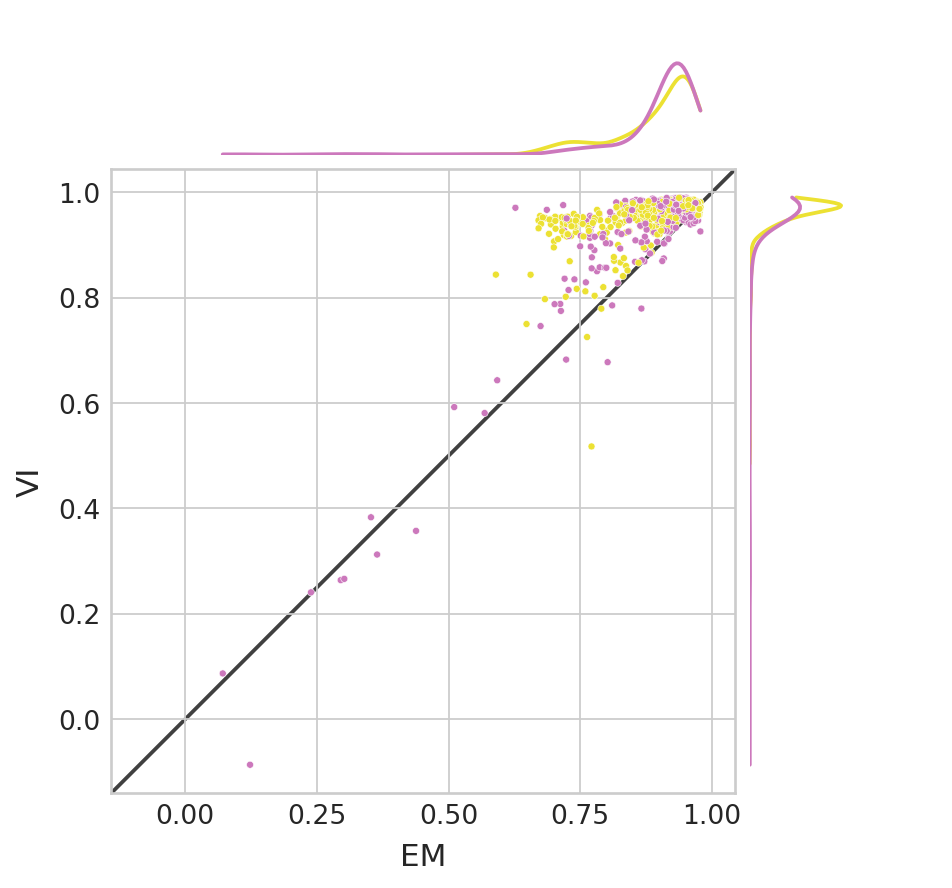
corr_df_velocity = pd.DataFrame(columns=["EM", "VI"], index=np.arange(len(adata.var_names)))
for var_id in range(len(adata.var_names)):
x = adata.layers["velocity"][:, var_id]
y = adata.layers["true_velocity"][:, var_id]
corr_df_velocity.loc[var_id, "EM"] = pearsonr(x, y)[0]
x = bdata.layers["velocity"][:, var_id]
corr_df_velocity.loc[var_id, "VI"] = pearsonr(x, y)[0]
corr_df_velocity['model'] = adata.var['model'].values
(corr_df_velocity["VI"] > corr_df_velocity["EM"]).mean()
0.9376163873370578
with mplscience.style_context():
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(figsize=(6, 6))
ax.axis('off')
left, width = 0.1, 0.65
bottom, height = 0.1, 0.65
spacing = 0.015
rect_scatter = [left, bottom, width, height]
rect_histx = [left, bottom + height + spacing, width, 0.1]
rect_histy = [left + width + spacing, bottom, 0.1, height]
ax = fig.add_axes(rect_scatter)
ax_histx = fig.add_axes(rect_histx, sharex=ax)
ax_histy = fig.add_axes(rect_histy, sharey=ax)
ax_histx.axis("off")
ax_histy.axis("off")
clipy = (corr_df_velocity["VI"].min(), corr_df_velocity["VI"].max())
clipx = (corr_df_velocity["EM"].min(), corr_df_velocity["EM"].max())
palette = dict(
zip(
["EM", "VI"],
[
sns.color_palette("colorblind").as_hex()[4],
sns.color_palette("colorblind").as_hex()[8]
]
)
)
sns.kdeplot(
data=corr_df_velocity,
y="VI",
hue="model",
ax=ax_histy,
legend=False,
clip=clipy,
palette=palette,
)
sns.kdeplot(
data=corr_df_velocity,
x="EM",
hue="model",
ax=ax_histx,
legend=False,
clip=clipx,
palette=palette,
)
sns.scatterplot(data=corr_df_velocity, x="EM", y="VI", hue='model', s=10, ax=ax, palette=palette, legend=False)
lims = [
np.min([ax.get_xlim(), ax.get_ylim()]), # min of both axes
np.max([ax.get_xlim(), ax.get_ylim()]), # max of both axes
]
# now plot both limits against eachother
ax.plot(lims, lims, 'k-', alpha=0.75, zorder=0)
ax.set_aspect('equal')
ax.set_xlim(lims)
ax.set_ylim(lims)
if SAVE_FIGURES:
fig.savefig(
FIG_DIR / 'comparison' / 'simulated_velo_correlation.svg',
format="svg",
transparent=True,
bbox_inches='tight'
)
ttest_res = ttest_ind(
corr_df_velocity["VI"].values.astype(float),
corr_df_velocity["EM"].values.astype(float),
equal_var=False
)
ttest_res
Ttest_indResult(statistic=15.858217619574068, pvalue=1.4333381236707924e-53)
if ttest_res.pvalue < 0.001:
significance = "***"
elif ttest_res.pvalue < 0.01:
significance = "**"
elif ttest_res.pvalue < 0.1:
significance = "*"
def add_significance(ax, left: int, right: int, significance: str, level: int = 0, **kwargs):
bracket_level = kwargs.pop("bracket_level", 1)
bracket_height = kwargs.pop("bracket_height", 0.02)
text_height = kwargs.pop("text_height", 0.01)
bottom, top = ax.get_ylim()
y_axis_range = top - bottom
bracket_level = (y_axis_range * 0.07 * level) + top * bracket_level
bracket_height = bracket_level - (y_axis_range * bracket_height)
ax.plot(
[left, left, right, right],
[bracket_height, bracket_level, bracket_level, bracket_height], **kwargs
)
ax.text(
(left + right) * 0.5,
bracket_level + (y_axis_range * text_height),
significance,
ha='center',
va='bottom',
c='k'
)
df = pd.melt(corr_df_velocity[["EM", "VI"]]).rename(columns={"variable": "Model", "value": "Velocity correlation"})
palette = dict(zip(["EM", "VI"], sns.color_palette("colorblind").as_hex()[:2]))
with mplscience.style_context():
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(figsize=(6, 4))
sns.boxplot(data=df, x="Model", y="Velocity correlation", palette=palette, ax=ax)
add_significance(
ax=ax, left=0, right=1, significance=significance, lw=1, c="k"
)
ylim_lower, ylim_upper = ax.get_ylim()
ax.set_ylim([ylim_lower, ylim_upper * 1.03])
ax.set_yticks([0, 0.25, 0.5, 0.75, 1])
if SAVE_FIGURES:
fig.savefig(
FIG_DIR / 'comparison' / 'simulated_velo_correlation_boxplot.svg',
format="svg",
transparent=True,
bbox_inches='tight'
)
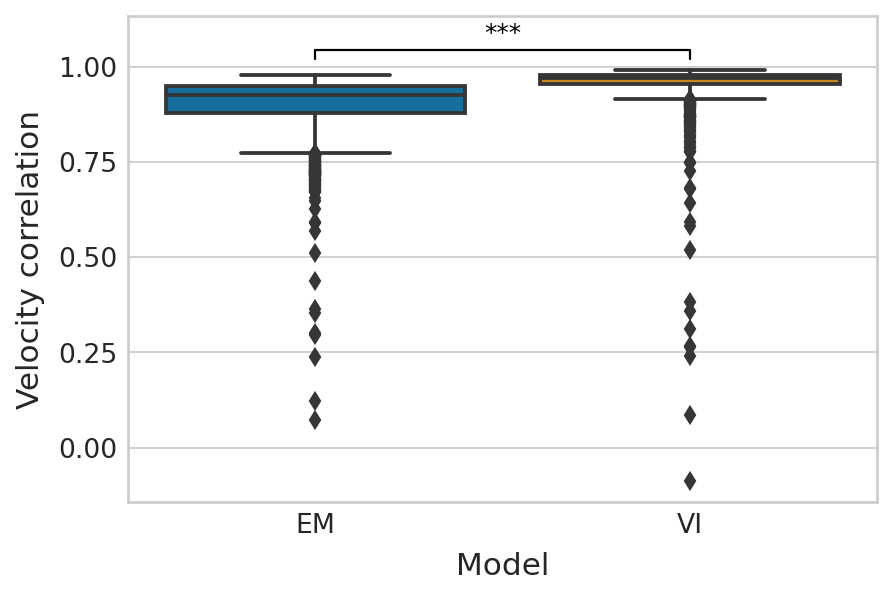
df = pd.melt(corr_df_velocity[["EM", "VI"]]).rename(columns={"variable": "Model", "value": "Velocity correlation"})
palette = dict(zip(["EM", "VI"], sns.color_palette("colorblind").as_hex()[:2]))
with mplscience.style_context():
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(figsize=(6, 4))
sns.boxplot(data=df, x="Model", y="Velocity correlation", palette=palette, ax=ax)
add_significance(
ax=ax,
left=0,
right=1,
significance=significance,
bracket_level=0.98,
bracket_height=0.01,
text_height=0.005,
lw=1,
c="k",
)
ax.set_ylim([0.75, 1.05])
if SAVE_FIGURES:
fig.savefig(
FIG_DIR / 'comparison' / 'simulated_velo_correlation_boxplot_clipped.svg',
format="svg",
transparent=True,
bbox_inches='tight'
)
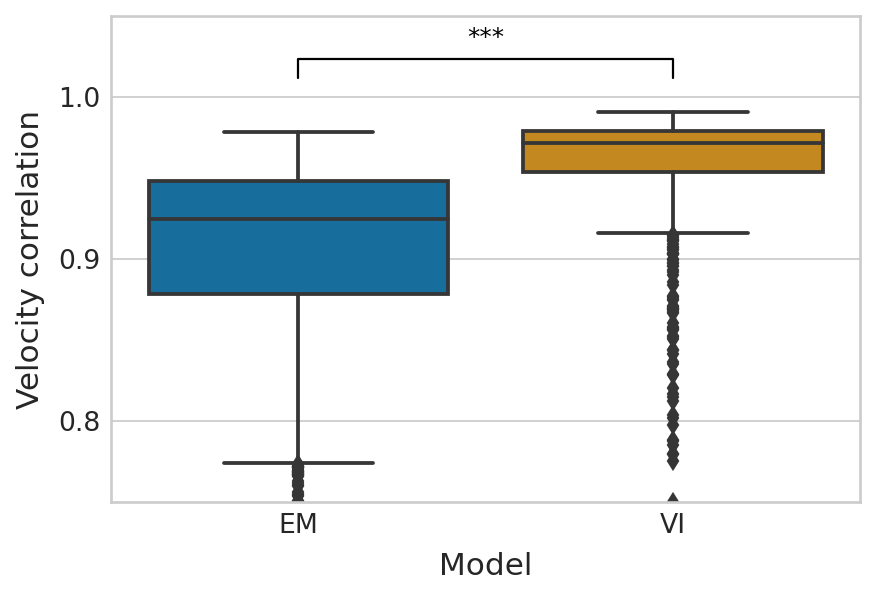
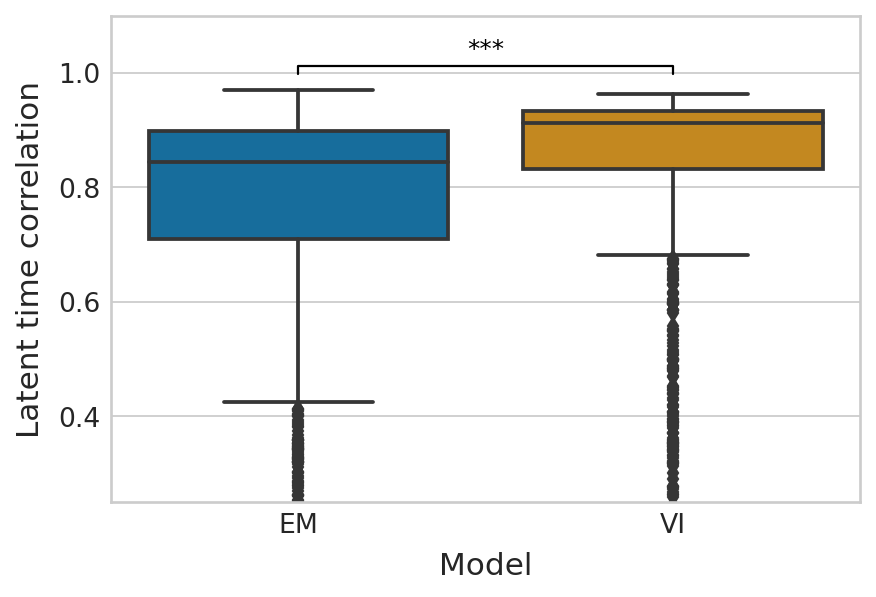
Latent time#
corr_df_time = pd.DataFrame(columns=["EM", "VI"], index=np.arange(len(adata.var_names)))
for var_id in range(len(adata.var_names)):
x = adata.layers["fit_t"][:, var_id]
y = adata.obs["true_t"]
corr_df_time.loc[var_id, "EM"] = pearsonr(x, y)[0]
x = bdata.layers["fit_t"][:, var_id]
corr_df_time.loc[var_id, "VI"] = pearsonr(x, y)[0]
corr_df_time['model'] = adata.var['model'].values
(corr_df_time["VI"] > corr_df_time["EM"]).mean()
0.8202979515828678
ttest_res = ttest_ind(
corr_df_time["VI"].values.astype(float),
corr_df_time["EM"].values.astype(float),
equal_var=False
)
ttest_res
Ttest_indResult(statistic=5.410308290240931, pvalue=6.996398279647197e-08)
if ttest_res.pvalue < 0.001:
significance = "***"
elif ttest_res.pvalue < 0.01:
significance = "**"
elif ttest_res.pvalue < 0.1:
significance = "*"
with mplscience.style_context():
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(figsize=(6, 6))
ax.axis('off')
left, width = 0.1, 0.65
bottom, height = 0.1, 0.65
spacing = 0.015
rect_scatter = [left, bottom, width, height]
rect_histx = [left, bottom + height + spacing, width, 0.1]
rect_histy = [left + width + spacing, bottom, 0.1, height]
ax = fig.add_axes(rect_scatter)
ax_histx = fig.add_axes(rect_histx, sharex=ax)
ax_histy = fig.add_axes(rect_histy, sharey=ax)
ax_histx.axis("off")
ax_histy.axis("off")
clipy = (corr_df_time["VI"].min(), corr_df_time["VI"].max())
clipx = (corr_df_time["EM"].min(), corr_df_time["EM"].max())
palette = dict(
zip(
["EM", "VI"],
[
sns.color_palette("colorblind").as_hex()[4],
sns.color_palette("colorblind").as_hex()[8]
]
)
)
sns.kdeplot(
data=corr_df_time,
y="VI",
hue="model",
ax=ax_histy,
legend=False,
clip=clipy,
palette=palette,
)
sns.kdeplot(
data=corr_df_time,
x="EM",
hue="model",
ax=ax_histx,
legend=False,
clip=clipx,
palette=palette,
)
sns.scatterplot(data=corr_df_time, x="EM", y="VI", hue='model', s=10, ax=ax, palette=palette, legend=False)
lims = [
np.min([ax.get_xlim(), ax.get_ylim()]), # min of both axes
np.max([ax.get_xlim(), ax.get_ylim()]), # max of both axes
]
# now plot both limits against eachother
ax.plot(lims, lims, 'k-', alpha=0.75, zorder=0)
ax.set_aspect('equal')
ax.set_xlim(lims)
ax.set_ylim(lims)
if SAVE_FIGURES:
fig.savefig(
FIG_DIR / 'comparison' / 'simulated_time_correlation.svg',
format="svg",
transparent=True,
bbox_inches='tight'
)
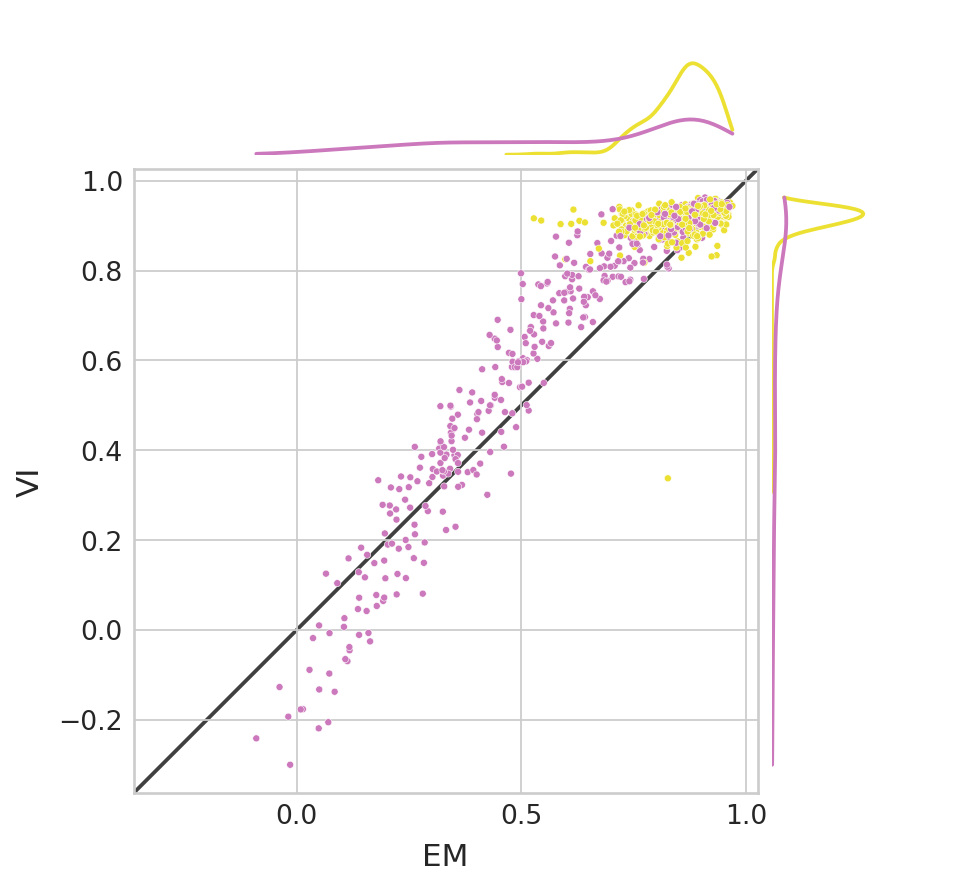
df = pd.melt(corr_df_time[["EM", "VI"]]).rename(columns={"variable": "Model", "value": "Latent time correlation"})
palette = dict(zip(["EM", "VI"], sns.color_palette("colorblind").as_hex()[:2]))
with mplscience.style_context():
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(figsize=(6, 4))
sns.boxplot(data=df, x="Model", y="Latent time correlation", palette=palette, ax=ax)
add_significance(
ax=ax, left=0, right=1, significance=significance, lw=1, c="k"
)
ylim_lower, ylim_upper = ax.get_ylim()
ax.set_ylim([ylim_lower, ylim_upper * 1.03])
ax.set_yticks([-0.25, 0, 0.25, 0.5, 0.75, 1])
if SAVE_FIGURES:
fig.savefig(
FIG_DIR / 'comparison' / 'simulated_time_correlation_boxplot.svg',
format="svg",
transparent=True,
bbox_inches='tight'
)
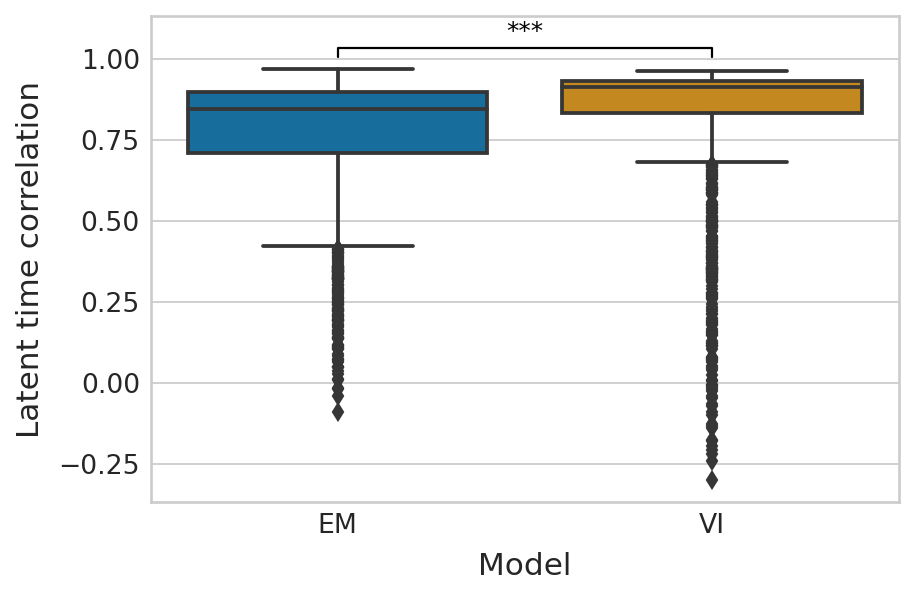
df = pd.melt(corr_df_time[["EM", "VI"]]).rename(columns={"variable": "Model", "value": "Latent time correlation"})
palette = dict(zip(["EM", "VI"], sns.color_palette("colorblind").as_hex()[:2]))
with mplscience.style_context():
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(figsize=(6, 4))
sns.boxplot(data=df, x="Model", y="Latent time correlation", palette=palette, ax=ax)
add_significance(
ax=ax,
left=0,
right=1,
significance=significance,
bracket_level=0.98,
bracket_height=0.01,
text_height=0.005,
lw=1,
c="k",
)
ax.set_ylim([0.25, 1.1])
if SAVE_FIGURES:
fig.savefig(
FIG_DIR / 'comparison' / 'simulated_time_correlation_boxplot_clipped.svg',
format="svg",
transparent=True,
bbox_inches='tight'
)