Comparison of velocities between EM and VI model#
Library imports#
%load_ext autoreload
%autoreload 2
import os
import sys
from pathlib import Path
from tqdm import tqdm
import numpy as np
import pandas as pd
import torch
from pygam import GAM
from scipy.stats import pearsonr, spearmanr, ttest_ind
from sklearn.metrics import accuracy_score
from velovi import preprocess_data, VELOVI
import matplotlib.pyplot as plt
import mplscience
import seaborn as sns
from matplotlib.colors import to_hex
import scanpy as sc
import scvelo as scv
import scvi
from scvelo.plotting.simulation import compute_dynamics
sys.path.append("../..")
from paths import DATA_DIR, FIG_DIR
from utils.gam import get_gams
Global seed set to 0
General settings#
scvi.settings.dl_pin_memory_gpu_training = False
sns.reset_defaults()
sns.reset_orig()
scv.settings.set_figure_params('scvelo', dpi_save=400, dpi=80, transparent=True, fontsize=20, color_map='viridis')
SAVE_FIGURES = True
if SAVE_FIGURES:
os.makedirs(FIG_DIR / 'comparison', exist_ok=True)
Function definitions#
def fit_scvelo(adata):
scv.tl.recover_dynamics(
adata, fit_scaling=False, var_names=adata.var_names, n_jobs=8
)
adata.var["fit_scaling"] = 1.0
scv.tl.velocity(adata, mode="dynamical", min_likelihood=-np.inf, min_r2=None)
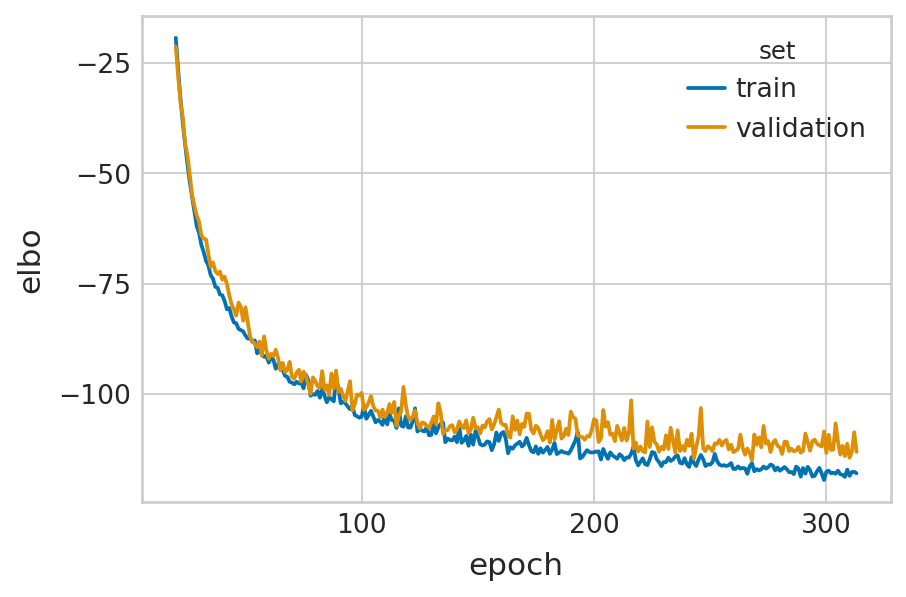
def fit_velovi(bdata):
VELOVI.setup_anndata(bdata, spliced_layer="Ms", unspliced_layer="Mu")
vae = VELOVI(bdata)
vae.train()
df = vae.history["elbo_train"].iloc[20:].reset_index().rename(columns={'elbo_train': 'elbo'})
df['set'] = 'train'
_df = vae.history["elbo_validation"].iloc[20:].reset_index().rename(columns={'elbo_validation': 'elbo'})
_df['set'] = 'validation'
df = pd.concat([df, _df], axis=0).reset_index(drop=True)
with mplscience.style_context():
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(figsize=(6, 4))
sns.lineplot(data=df, x='epoch', y='elbo', hue='set', palette=['#0173B2', '#DE8F05'], ax=ax)
latent_time = vae.get_latent_time(n_samples=25)
velocities = vae.get_velocity(n_samples=25, velo_statistic="mean")
t = latent_time
scaling = 20 / t.max(0)
bdata.layers["velocities_velovi"] = velocities / scaling
bdata.layers["latent_time_velovi"] = latent_time
bdata.var["fit_alpha"] = vae.get_rates()["alpha"] / scaling
bdata.var["fit_beta"] = vae.get_rates()["beta"] / scaling
bdata.var["fit_gamma"] = vae.get_rates()["gamma"] / scaling
bdata.var["fit_t_"] = (
torch.nn.functional.softplus(vae.module.switch_time_unconstr)
.detach()
.cpu()
.numpy()
) * scaling
bdata.layers["fit_t"] = latent_time.values * scaling[np.newaxis, :]
bdata.var['fit_scaling'] = 1.0
return vae
def compute_confidence(adata, vkey="velocity"):
velo = adata.layers[vkey]
scv.tl.velocity_graph(adata, vkey=vkey, n_jobs=8)
scv.tl.velocity_confidence(adata, vkey=vkey)
g_df = pd.DataFrame()
g_df["Velocity confidence"] = adata.obs[f"{vkey}_confidence"].to_numpy().ravel()
return g_df
def add_significance(ax, left: int, right: int, significance: str, level: int = 0, **kwargs):
bracket_level = kwargs.pop("bracket_level", 1)
bracket_height = kwargs.pop("bracket_height", 0.02)
text_height = kwargs.pop("text_height", 0.01)
bottom, top = ax.get_ylim()
y_axis_range = top - bottom
bracket_level = (y_axis_range * 0.07 * level) + top * bracket_level
bracket_height = bracket_level - (y_axis_range * bracket_height)
ax.plot(
[left, left, right, right],
[bracket_height, bracket_level, bracket_level, bracket_height], **kwargs
)
ax.text(
(left + right) * 0.5,
bracket_level + (y_axis_range * text_height),
significance,
ha='center',
va='bottom',
c='k'
)
def get_classification_scores(velo_sign_true, velo_sign_pred, score_fun, **kwargs):
n_vars = velo_sign_true.shape[1]
em_score = [
score_fun(velo_sign_true[:, var_id], velo_sign_pred["EM"][:, var_id], **kwargs)
for var_id in range(n_vars)
]
vi_score = [
score_fun(velo_sign_true[:, var_id], velo_sign_pred["VI"][:, var_id], **kwargs)
for var_id in range(n_vars)
]
return em_score, vi_score
def get_significance(pvalue):
if pvalue < 0.001:
return "***"
elif pvalue < 0.01:
return "**"
elif pvalue < 0.1:
return "*"
else:
return "n.s."
def plot_classification_score(em_score, vi_score, baseline_performance, score_name, **kwargs):
n_vars = len(em_score)
df = pd.DataFrame(
{
score_name: vi_score + em_score + baseline_performance,
"Model": ["VI"] * n_vars + ["EM"] * n_vars + ["Random"] * n_vars
}
)
with mplscience.style_context():
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(figsize=(6, 4))
sns.boxplot(data=df, x="Model", y=score_name, palette="colorblind", ax=ax)
Data loading#
adata = sc.read(DATA_DIR / "scEU" / "gex_raw.h5ad")
adata = adata[adata.obs["labeling_time"] != "dmso", :]
adata.obs['labeling_time'] = adata.obs['labeling_time'].astype(float)
adata = adata[adata.obs['experiment'] == "Pulse", :].copy()
adata.layers["unspliced"] = adata.layers["unlabeled_unspliced"] + adata.layers["labeled_unspliced"]
adata.layers["spliced"] = adata.layers["unlabeled_spliced"] + adata.layers["labeled_spliced"]
adata.obs["pseudo_clusters"] = pd.cut(adata.obs["cell_cycle_position"], bins=30, labels=False).astype(str).astype("category")
adata.obs["pseudo_clusters_equal_size"] = pd.qcut(adata.obs["cell_cycle_position"], q=30, labels=False)
adata.obs["pseudo_clusters_equal_size_num"] = adata.obs["pseudo_clusters_equal_size"].astype(float)
adata.obs["cell_cycle_rad"] = adata.obs["cell_cycle_position"] / adata.obs["cell_cycle_position"].max() * 2 * np.pi
adata
AnnData object with n_obs × n_vars = 2793 × 11848
obs: 'experiment', 'labeling_time', 'plate_id', 'well_id', 'cell_cycle_position', 'RFP_log10_corrected', 'GFP_log10_corrected', 'pseudo_clusters', 'pseudo_clusters_equal_size', 'pseudo_clusters_equal_size_num', 'cell_cycle_rad'
layers: 'labeled_spliced', 'labeled_unspliced', 'new', 'total', 'unlabeled_spliced', 'unlabeled_unspliced', 'unspliced', 'spliced'
Data preprocessing#
scv.pp.filter_and_normalize(adata, min_counts=10, layers_normalize=['X', 'new', 'total', 'unspliced', 'spliced'], n_top_genes=2000)
Filtered out 905 genes that are detected 10 counts (spliced).
Normalized count data: X, new, total, unspliced, spliced.
Extracted 2000 highly variable genes.
Logarithmized X.
sc.tl.pca(adata, svd_solver='arpack')
sc.pp.neighbors(adata, n_neighbors=30, n_pcs=30)
scv.pp.moments(adata, n_pcs=None, n_neighbors=None)
adata.layers["Ms_log1p"] = np.log1p(adata.layers["Ms"])
computing moments based on connectivities
finished (0:00:00) --> added
'Ms' and 'Mu', moments of un/spliced abundances (adata.layers)
sc.tl.umap(adata)
with mplscience.style_context():
fig, ax = plt.subplots(figsize=(6, 4))
scv.pl.scatter(adata, basis='umap', color='cell_cycle_position', cmap='viridis', title='', ax=ax)
if SAVE_FIGURES:
fig.savefig(
FIG_DIR / 'comparison' / 'sceu_cell_cycle_umap.svg',
format="svg",
transparent=True,
bbox_inches='tight'
)

adata = preprocess_data(adata)
bdata = adata.copy()
cdata = adata.copy()
computing velocities
finished (0:00:00) --> added
'velocity', velocity vectors for each individual cell (adata.layers)
((adata.layers["Mu"] == 0) & (adata.layers["Ms"] == 0)).mean()
0.008771929824561403
Model fitting#
EM model#
fit_scvelo(adata)
recovering dynamics (using 8/14 cores)
Global seed set to 0
Global seed set to 0
Global seed set to 0
Global seed set to 0
Global seed set to 0
Global seed set to 0
Global seed set to 0
Global seed set to 0
finished (0:00:08) --> added
'fit_pars', fitted parameters for splicing dynamics (adata.var)
computing velocities
finished (0:00:00) --> added
'velocity', velocity vectors for each individual cell (adata.layers)
VI model#
velovi_vae = fit_velovi(bdata)
GPU available: True (cuda), used: True
TPU available: False, using: 0 TPU cores
IPU available: False, using: 0 IPUs
HPU available: False, using: 0 HPUs
LOCAL_RANK: 0 - CUDA_VISIBLE_DEVICES: [0]
Epoch 314/500: 63%|██████▎ | 314/500 [01:59<01:10, 2.63it/s, loss=-121, v_num=1]
Monitored metric elbo_validation did not improve in the last 45 records. Best score: -115.002. Signaling Trainer to stop.
Empirical velocity#
aggr_counts = []
for pos in tqdm(np.sort(adata.obs["cell_cycle_position"].unique())):
mask = (adata.obs["cell_cycle_position"] == pos).values
aggr_counts.append(np.median(adata.layers["Ms"][mask, :], axis=0))
aggr_counts = np.vstack(aggr_counts)
100%|██████████| 290/290 [00:00<00:00, 4516.30it/s]
reorder_mask = np.arange(1, adata.obs["cell_cycle_rad"].nunique()).tolist() + [0]
cell_cycle_pos = adata.obs["cell_cycle_rad"].values
cc_pos_diff = np.sort(np.unique(cell_cycle_pos))
cc_pos_diff = (cc_pos_diff[reorder_mask] - cc_pos_diff) % (2 * np.pi)
empirical_velo = (aggr_counts[reorder_mask, :] - aggr_counts) / cc_pos_diff.reshape(-1, 1)
empirical_velo_sign = np.sign(empirical_velo)
Velocity processing#
aggr_velo = {"EM": [], "VI": []}
for pos in tqdm(np.sort(adata.obs["cell_cycle_position"].unique())):
mask = (adata.obs["cell_cycle_position"] == pos).values
aggr_velo["EM"].append(np.median(adata.layers["velocity"][mask, :], axis=0))
aggr_velo["VI"].append(np.median(bdata.layers["velocities_velovi"][mask, :], axis=0))
aggr_velo["EM"] = np.vstack(aggr_velo["EM"])
aggr_velo["VI"] = np.vstack(aggr_velo["VI"])
100%|██████████| 290/290 [00:00<00:00, 3295.36it/s]
aggr_velo_sign = {}
aggr_velo_sign["EM"] = np.sign(aggr_velo["EM"])
aggr_velo_sign["VI"] = np.sign(aggr_velo["VI"])
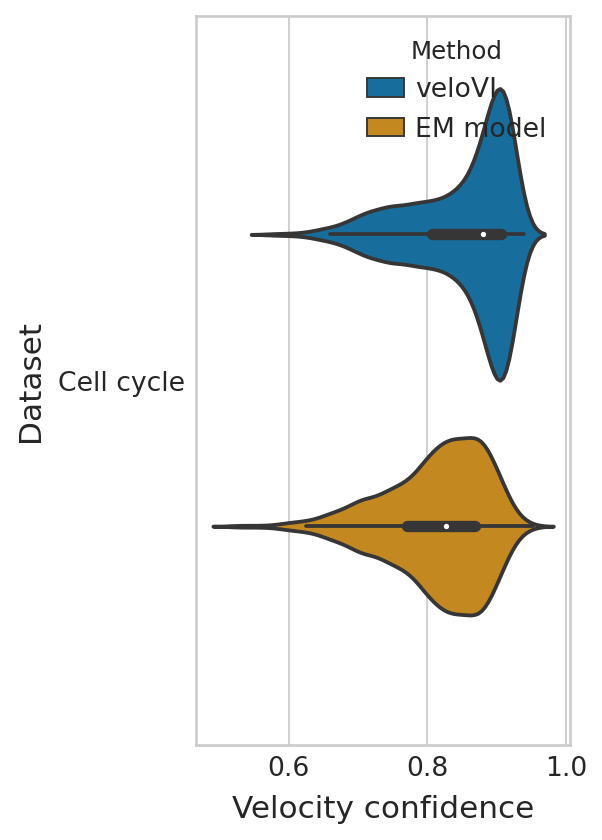
Velocity confidence#
dfs = []
g_df = compute_confidence(adata)
g_df["Dataset"] = "Cell cycle"
g_df["Method"] = "EM model"
dfs.append(g_df)
g_df = compute_confidence(bdata, vkey="velocities_velovi")
g_df["Dataset"] = "Cell cycle"
g_df["Method"] = "veloVI"
dfs.append(g_df)
conf_df = pd.concat(dfs, axis=0)
computing velocity graph (using 8/14 cores)
finished (0:00:02) --> added
'velocity_graph', sparse matrix with cosine correlations (adata.uns)
--> added 'velocity_length' (adata.obs)
--> added 'velocity_confidence' (adata.obs)
--> added 'velocity_confidence_transition' (adata.obs)
computing velocity graph (using 8/14 cores)
finished (0:00:02) --> added
'velocities_velovi_graph', sparse matrix with cosine correlations (adata.uns)
--> added 'velocities_velovi_length' (adata.obs)
--> added 'velocities_velovi_confidence' (adata.obs)
--> added 'velocities_velovi_confidence_transition' (adata.obs)
with mplscience.style_context():
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(figsize=(3, 6))
sns.violinplot(
data=conf_df,
ax=ax,
orient="h",
y="Dataset",
x="Velocity confidence",
hue="Method",
hue_order=["veloVI", "EM model"],
palette=sns.color_palette(['#0173B2', '#DE8F05']),
);
if SAVE_FIGURES:
fig.savefig(
FIG_DIR / 'comparison' / 'sceu_velocity_confidence.svg',
format="svg",
transparent=True,
bbox_inches='tight'
)
/vol/storage/miniconda3/envs/velovi-py39/lib/python3.9/site-packages/seaborn/categorical.py:381: DeprecationWarning: distutils Version classes are deprecated. Use packaging.version instead.
if LooseVersion(mpl.__version__) < "3.0":
/vol/storage/miniconda3/envs/velovi-py39/lib/python3.9/site-packages/setuptools/_distutils/version.py:346: DeprecationWarning: distutils Version classes are deprecated. Use packaging.version instead.
other = LooseVersion(other)
GAM fitting#
gams = {}
gams["EM"] = get_gams(adata=adata, layer="velocity", time="cell_cycle_rad", n_jobs=8)
gams["VI"] = get_gams(adata=bdata, layer="velocities_velovi", time="cell_cycle_rad", n_jobs=8)
100%|██████████| 140/140 [00:00<00:00, 258.55it/s]
100%|██████████| 140/140 [00:00<00:00, 262.05it/s]
cell_cycle_score = adata.obs["cell_cycle_rad"].values
cell_cycle_score = np.vstack([cell_cycle_score - 2 * np.pi, cell_cycle_score, cell_cycle_score + 2 * np.pi]).T
adata.var["gam_r2_scores"] = [
gam._estimate_r2(X=cell_cycle_score, y=adata.layers["velocity"][:, var_id])["McFadden"]
for var_id, gam in enumerate(gams["EM"].values())
]
bdata.var["gam_r2_scores"] = [
gam._estimate_r2(X=cell_cycle_score, y=bdata.layers["velocities_velovi"][:, var_id])["McFadden"]
for var_id, gam in enumerate(gams["VI"].values())
]
/vol/storage/miniconda3/envs/velovi-py39/lib/python3.9/site-packages/pygam/utils.py:649: DeprecationWarning: `np.int` is a deprecated alias for the builtin `int`. To silence this warning, use `int` by itself. Doing this will not modify any behavior and is safe. When replacing `np.int`, you may wish to use e.g. `np.int64` or `np.int32` to specify the precision. If you wish to review your current use, check the release note link for additional information.
Deprecated in NumPy 1.20; for more details and guidance: https://numpy.org/devdocs/release/1.20.0-notes.html#deprecations
bases = (x >= aug_knots[:-1]).astype(np.int) * \
/vol/storage/miniconda3/envs/velovi-py39/lib/python3.9/site-packages/pygam/utils.py:650: DeprecationWarning: `np.int` is a deprecated alias for the builtin `int`. To silence this warning, use `int` by itself. Doing this will not modify any behavior and is safe. When replacing `np.int`, you may wish to use e.g. `np.int64` or `np.int32` to specify the precision. If you wish to review your current use, check the release note link for additional information.
Deprecated in NumPy 1.20; for more details and guidance: https://numpy.org/devdocs/release/1.20.0-notes.html#deprecations
(x < aug_knots[1:]).astype(np.int)
df = pd.DataFrame(
{
"EM": adata.var["gam_r2_scores"].values,
"VI": bdata.var["gam_r2_scores"].values,
}
)
with mplscience.style_context():
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(figsize=(6, 6))
sns.scatterplot(data=df, x="EM", y="VI", ax=ax)
lims = [
np.min([ax.get_xlim(), ax.get_ylim()]), # min of both axes
np.max([ax.get_xlim(), ax.get_ylim()]), # max of both axes
]
# now plot both limits against eachother
ax.plot(lims, lims, 'k-', alpha=0.75, zorder=0)
ax.set_aspect('equal')
ax.set_xlim(lims)
ax.set_ylim(lims);
if SAVE_FIGURES:
fig.savefig(
FIG_DIR / 'comparison' / 'sceu_gam_r2.svg',
format="svg",
transparent=True,
bbox_inches='tight'
)
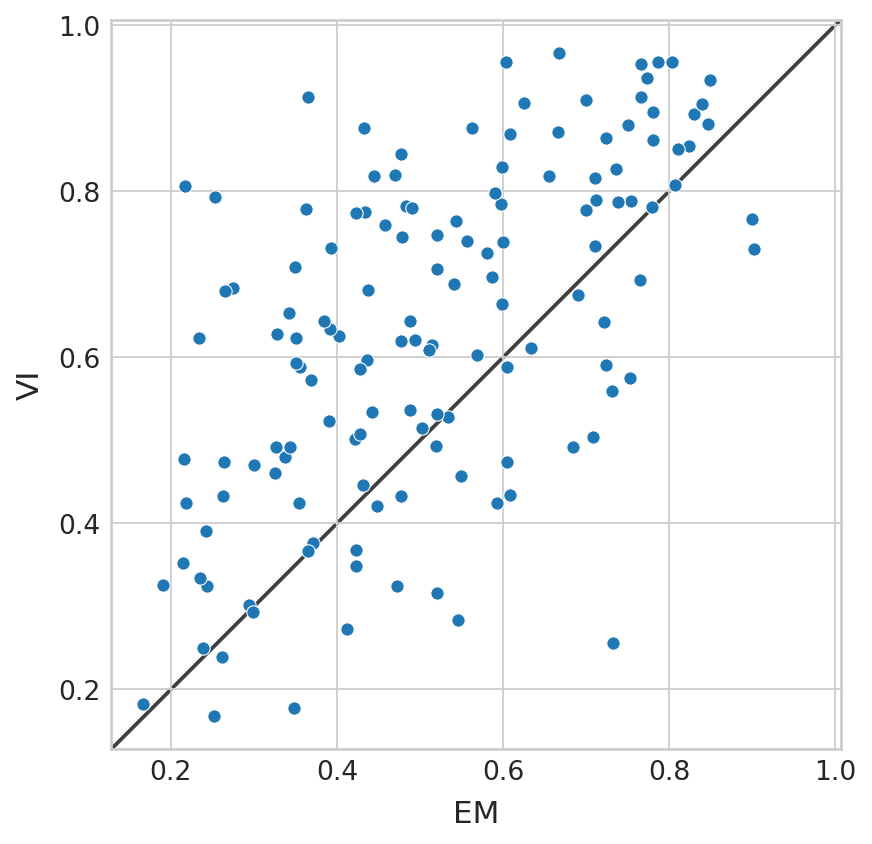
(bdata.var["gam_r2_scores"].values > adata.var["gam_r2_scores"].values).mean()
0.7785714285714286
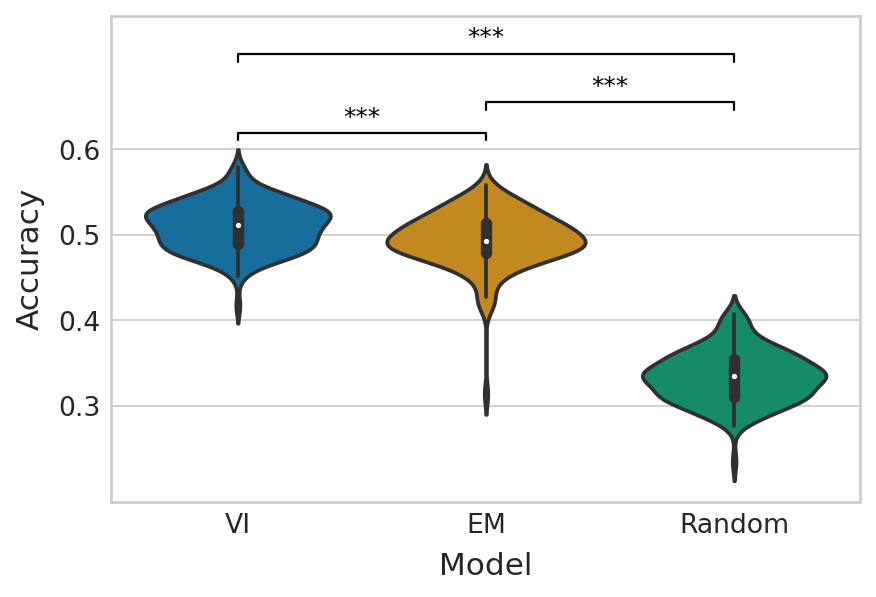
Estimated velocity vs. empirical velocity#
np.random.seed(0)
random_velo_sign = np.random.choice([-1, 0, 1], size=(len(adata.obs["cell_cycle_position"].unique()), adata.n_vars))
baseline_performance = [
accuracy_score(empirical_velo_sign[:, var_id], random_velo_sign[:, var_id])
for var_id in range(adata.n_vars)
]
em_score, vi_score = get_classification_scores(
velo_sign_true=empirical_velo_sign,
velo_sign_pred=aggr_velo_sign,
score_fun=accuracy_score,
)
df = pd.DataFrame(
{
"Accuracy": vi_score + em_score + baseline_performance,
"Model": ["VI"] * adata.n_vars + ["EM"] * adata.n_vars + ["Random"] * adata.n_vars
}
)
with mplscience.style_context():
sns.set_style(style="whitegrid")
fig, ax = plt.subplots(figsize=(6, 4))
sns.violinplot(data=df, x="Model", y="Accuracy", palette="colorblind", ax=ax)
ttest_res = ttest_ind(vi_score, em_score, equal_var=False, alternative="greater")
significance = get_significance(ttest_res.pvalue)
add_significance(
ax=ax, left=0, right=1, significance=significance, lw=1, c="k", level=0,
)
ttest_res = ttest_ind(em_score, baseline_performance, equal_var=False, alternative="greater")
significance = get_significance(ttest_res.pvalue)
add_significance(
ax=ax, left=1, right=2, significance=significance, lw=1, c="k", level=1, bracket_level=0.975
)
ttest_res = ttest_ind(em_score, baseline_performance, equal_var=False, alternative="greater")
significance = get_significance(ttest_res.pvalue)
add_significance(
ax=ax, left=0, right=2, significance=significance, lw=1, c="k", level=2, bracket_level=0.95
)
y_min, y_max = ax.get_ylim()
ax.set_ylim([y_min, y_max + 0.02])
ax.set_yticks([0.3, 0.4, 0.5, 0.6])
ax.set_yticklabels([0.3, 0.4, 0.5, 0.6]);
if SAVE_FIGURES:
fig.savefig(
FIG_DIR / 'comparison' / 'sceu_velocity_sign_accuracy.svg',
format="svg",
transparent=True,
bbox_inches='tight'
)