Tutorial¶
Contents
Broadly speaking, Compass takes in a gene expression matrix scaled for library depth (e.g., CPM), and outputs a penalty reaction matrix, whereby higher scores correspond to a reaction being less likely.
Running Compass (Simple)¶
The input gene expression matrix can be either a tab-delimited text file (tsv) or a matrix market format (mtx) containing gene expression estimates (CPM, TPM, or similar scaled units) with one row per gene, one column per sample.
Tab-delimited files need row and column labels corresponding to genes and sample names. Market matrix formats need a separate tab delimited file of of gene names and optionally a tab delimited file of cell names.
Example input¶
You can find example inputs in tab-delimited format (tsv) and market matrix format (mtx) on this github repo under compass/Resources/Test-Data.
These files will exist locally as well under the Compass install directory which can be found by running:
compass --example-inputs --species homo_sapiens
Human or mouse species makes no difference for this command.
Running Compass¶
Then after opening a command line in a directory with an input file “expression.tsv”, you can run Compass on the data with the following command, which will limit the number of processes to 10:
compass --data expression.tsv --num-processes 10 --species homo_sapiens
And to run Compass on mtx formatted data use the following:
compass --data-mtx expression.mtx genes.tsv sample_names.tsv --num-processes 10 --species homo_sapiens
Though the sample names file can be omitted, in which case the samples will be labelled by index.
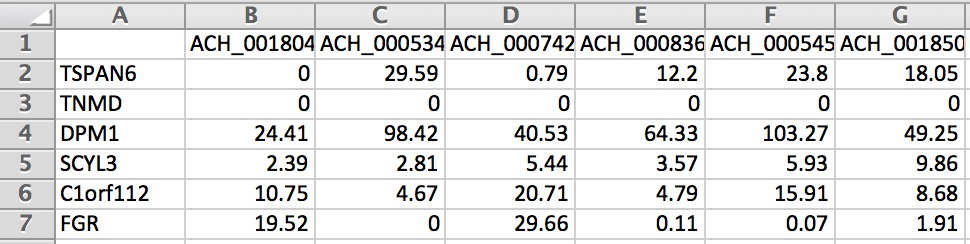
Below is an example of the formatting for gene expression (We only show a small portion of the matrix):
For the first run of Compass on a given model and media there will be overhead building up Compass’s cache. Compass will automatically build up the cache if it is empty, but you can also manually build up the cache before running Compass with:
compass --precache --species homo_sapiens
Note
For every individual sample, Compass takes roughly 30 minutes to calculate the reaction penalties (varying by machine). This can be expedited by running more than one process at once. In addition, Compass saves the results of all samples that it has already processed in the _tmp directory. Therefore, Compass can also be stopped and restarted after it is done processing a subset of samples so long as the _tmp directory is still there.
Compass Settings¶
Compass also allows users to customize a variety of settings seen below:
usage: Compass [-h] [--data FILE] [--data-mtx FILE [FILE ...]] [--model MODEL]
[--species SPECIES] [--media MEDIA] [--output-dir DIR]
[--temp-dir DIR] [--torque-queue QUEUE] [--num-processes N]
[--lambda F] [--num-threads N] [--and-function FXN]
[--select-reactions FILE] [--num-neighbors N]
[--symmetric-kernel] [--input-weights FILE]
[--penalty-diffusion MODE] [--no-reactions]
[--calc-metabolites] [--precache] [--input-knn FILE]
[--output-knn FILE] [--latent-space FILE] [--only-penalties]
[--list-genes FILE]
See the instructions here for an in depth tutorial on using Compass’s settings
Micropooling¶
The Compass algorithm can be very computationally intensive, especially for large datasets, but with micropooling/clustering techniques you can reduce the computing time by an order of magnitude. Compass comes with a micropooling algorithm built in, based on a reimplementation of microclustering from VISION. To enable microclustering you can specify a microcluster size as below:
Compass --microcluster-size 10 [other options]
In general cluster size presents a tradeoff between runtime and granularity as larger clusters can make analysis more sensitive but will take longer to process the samples, so a microcluster size as small as computationally feasible is recommended. There are more details on micropooling with Compass here.
Alternatively, any other method of aggregating cells into fewer representatives can be used such as metacell.
Postprocessing¶
Once Compass has finished running, we apply several steps of postprocessing to the data. Mainly, the postprocessing converts reaction penalties (where high values correspond to low likelihood reactions) to reaction scores (where high values correspond to likely reactions).
Outputs¶
When Compass has completed, the outputs for all samples are stored in a
tab delimited file reactions.tsv in the specified output directory
(. directory when running Compass by default).
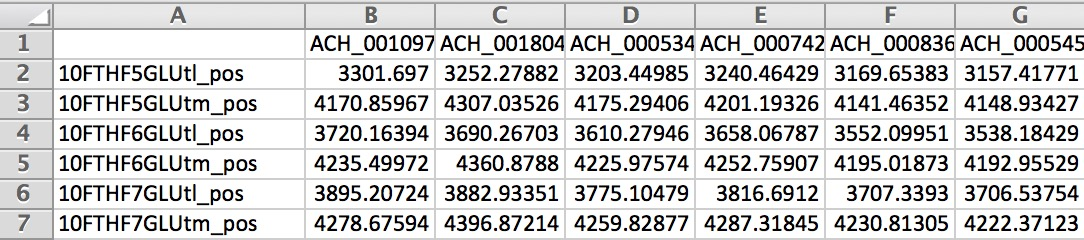
Below is an example of the output matrix:
To get more context on what the reaction identifiers are, you can visit virtual metabolic human or the resources directory of Compass where there are several csv’s which include information on the reactions in Recon2.
Note: While Compass is running, it will store partial results for each sample in the _tmp directory/ (or the directory following --temp-dir)